Research
Research

Ecology and Evolution of Large DNA Viruses
Large DNA viruses have emerged as important constituents of ecosystems around the globe. These “giant viruses” collectively play critical roles in the biosphere by influencing global biogeochemical cycles and shaping host genome evolution through endogenization. Their genomes are unusually complex and encode many genes typically found only in cellular lineages, such as those involved in the TCA cycle, glycolysis, amino acid metabolism, light sensing, sphingolipid biosynthesis, cytoskeletal structure, and fermentation. We use a variety of computational and wet-lab approaches to better understand the evolution, genomics, and host-virus interactions of these remarkable viruses.
Some related publications
FO Aylward, M Moniruzzaman, AD Ha, EV Koonin. A phylogenomic framework for charting the diversity and evolution of giant viruses. PLOS Biology, 2021, 19(10).
M Moniruzzaman, AR Weinheimer, CA Martinez-Gutierrez, FO Aylward. Widespread endogenization of giant viruses shapes genomes of green algae. Nature, 2020.
M Moniruzzaman, CA Martinez-Gutierrez, AR Weinheimer, FO Aylward. Dynamic genome evolution and complex virocell metabolism of globally-distributed giant viruses. Nature Communications, 2020, 11(1): 1-11.
AR Weinheimer & FO Aylward. Infection strategy and biogeography distinguish cosmopolitan groups of jumbo bacteriophages. The ISME Journal, 16: 1657–1667.
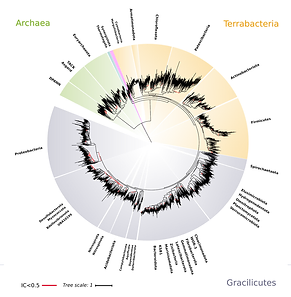
Microbial Diversity and the Tree of Life
We are interested in examining the ecology and evolution of major bacterial and archaeal lineages across the Tree of Life. This work includes broad phylogenomic analysis of evolutionary relationships across Bacteria and Archaea, as well as more directed analysis of the genome evolution of particular lineages. Many of the ancient lineages we examine have had a profound influence in shaping the chemical environment of Earth, yet their ecology and evolutionary history often remain obscure.
Some related publications
CA Martinez-Gutierrez, JC Uyeda, FO Aylward. A Timeline of Bacterial and Archaeal Diversification in the Ocean. BioRxiv, 2022. https://doi.org/10.1101/2022.10.27.514092
CA Martinez-Gutierrez & FO Aylward. Phylogenetic signal, congruence, and uncertainty across bacteria and archaea. Molecular Biology and Evolution, 2021, 38(12).
CA Martinez-Gutierrez & FO Aylward. Strong purifying selection is associated with genome streamlining in epipelagic Marinimicrobia. Genome Biology and Evolution, 2019, 11(10): 2887-2894.
EW Getz, SS Tithi, L Zhang, FO Aylward. Parallel evolution of key genomic features and cellular bioenergetics across the marine radiation of a bacterial phylum. mBio, 2018.
